Importing data
Data Hierarchy
Data within NiDB is stored in a hierarchy:
The top level data item is a subject.
Subjects are enrolled in projects
Each subject has imaging studies, associated with an enrollment
Each study has series
Each series has files
See diagrams and examples of the hierarchy here.
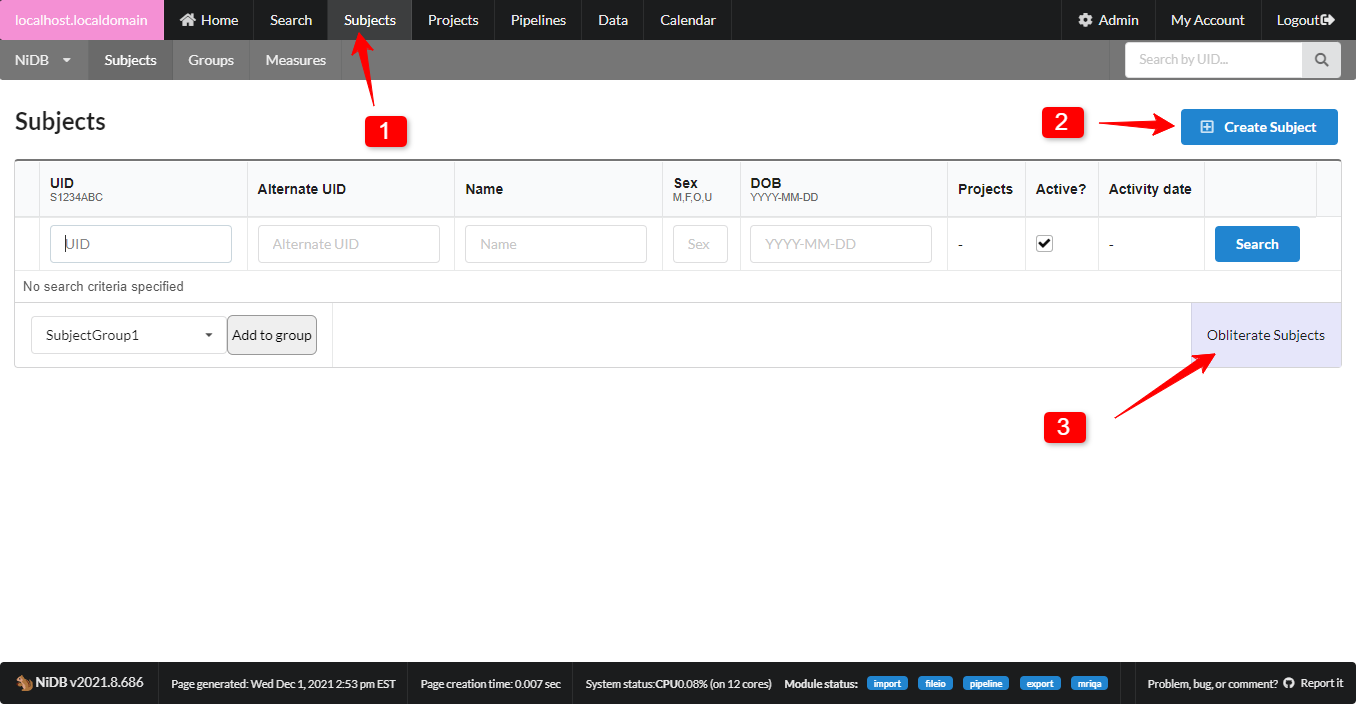
Create Subject
On the main menu, find the Subjects tab. A page will be displayed in which you can search for existing subjects, and a button to create a new subject
Subjects page menu item
Create Subject button
Obliterate subjects button: an intimidating sounding button that only appears for NiDB admins
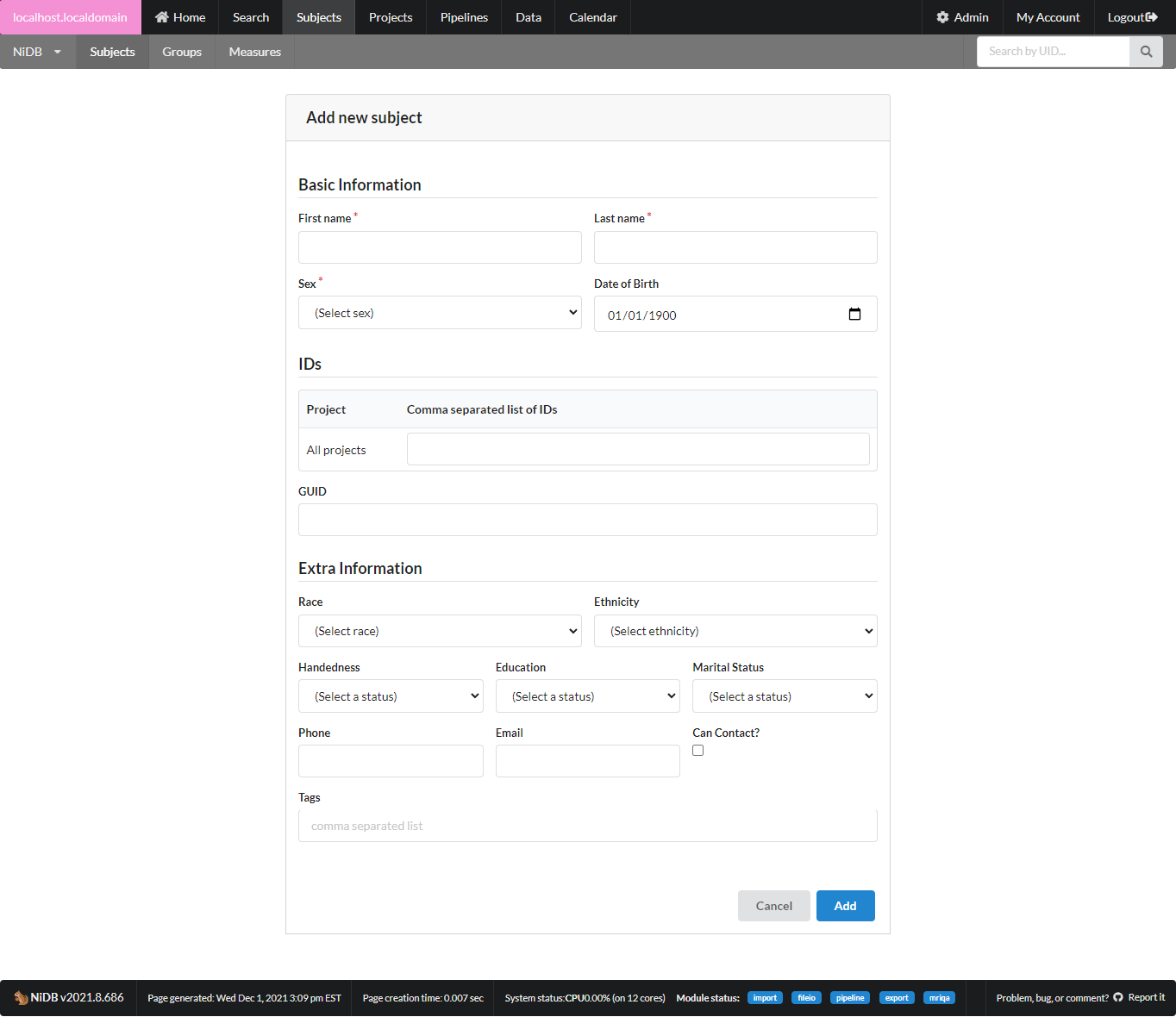
Fill out as much information as you need. Name, Sex, DOB are required to ensure a unique subject. Most other information is optional. While fields for contact information are available, be mindful and consider whether you really need to fill those out. Chances are that contact information for research participants is already stored in a more temporary location and does not need to exist for as long as the imaging data does.
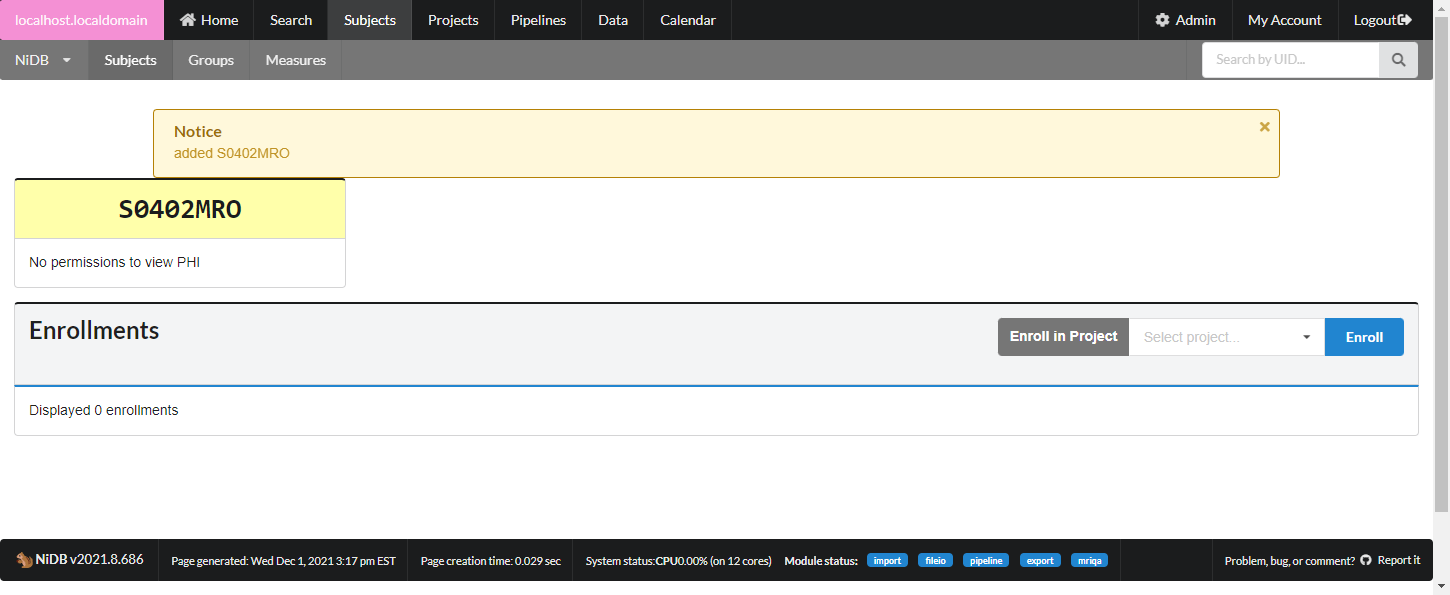
The subject will now be assigned a UID, but will not be enrolled in any projects. Enroll the subject in the next section.
Importing Subjects from Recap
For a project, subjects can be imported from redcap using an option on the project page as shown below:
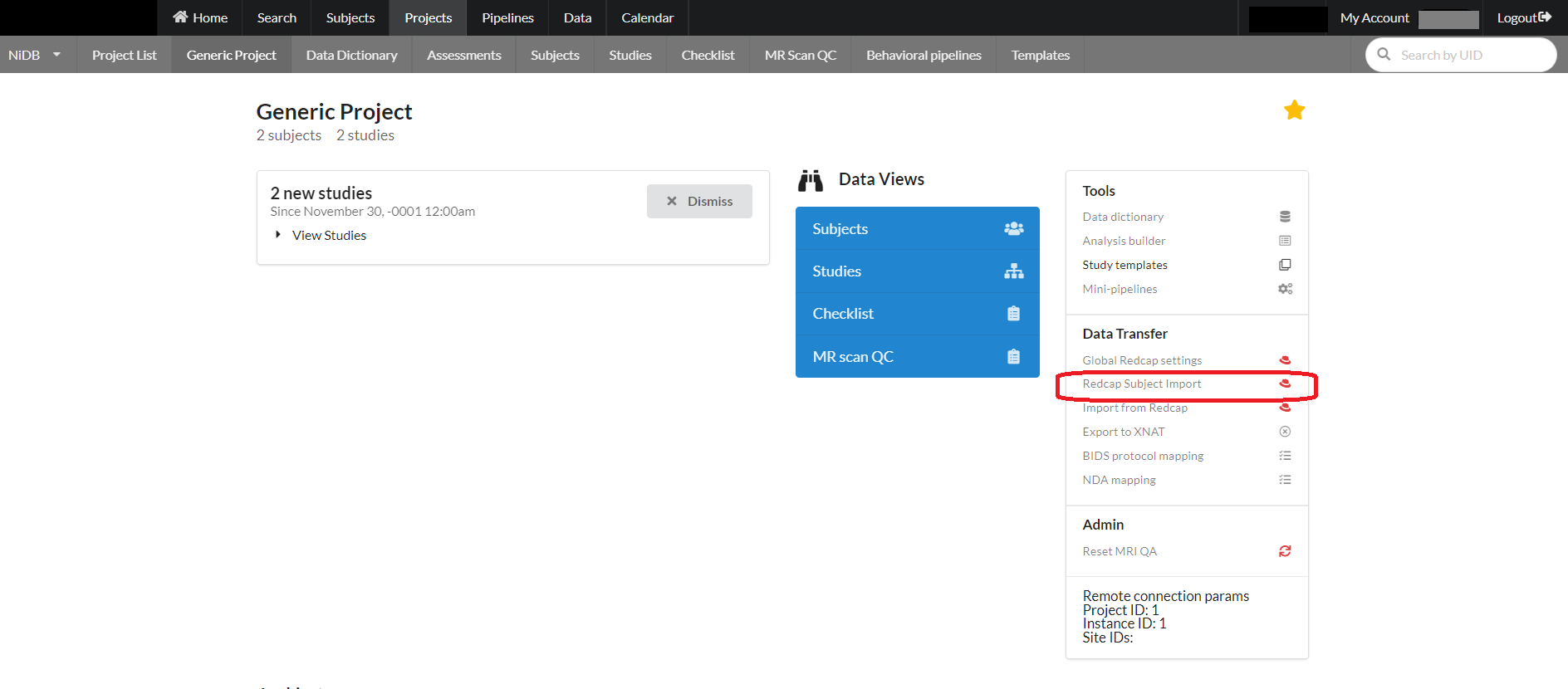
Fill the following form requiring information for API connection to redcap and required redcap field names. After providing the required fields click "Subjects Information" button.
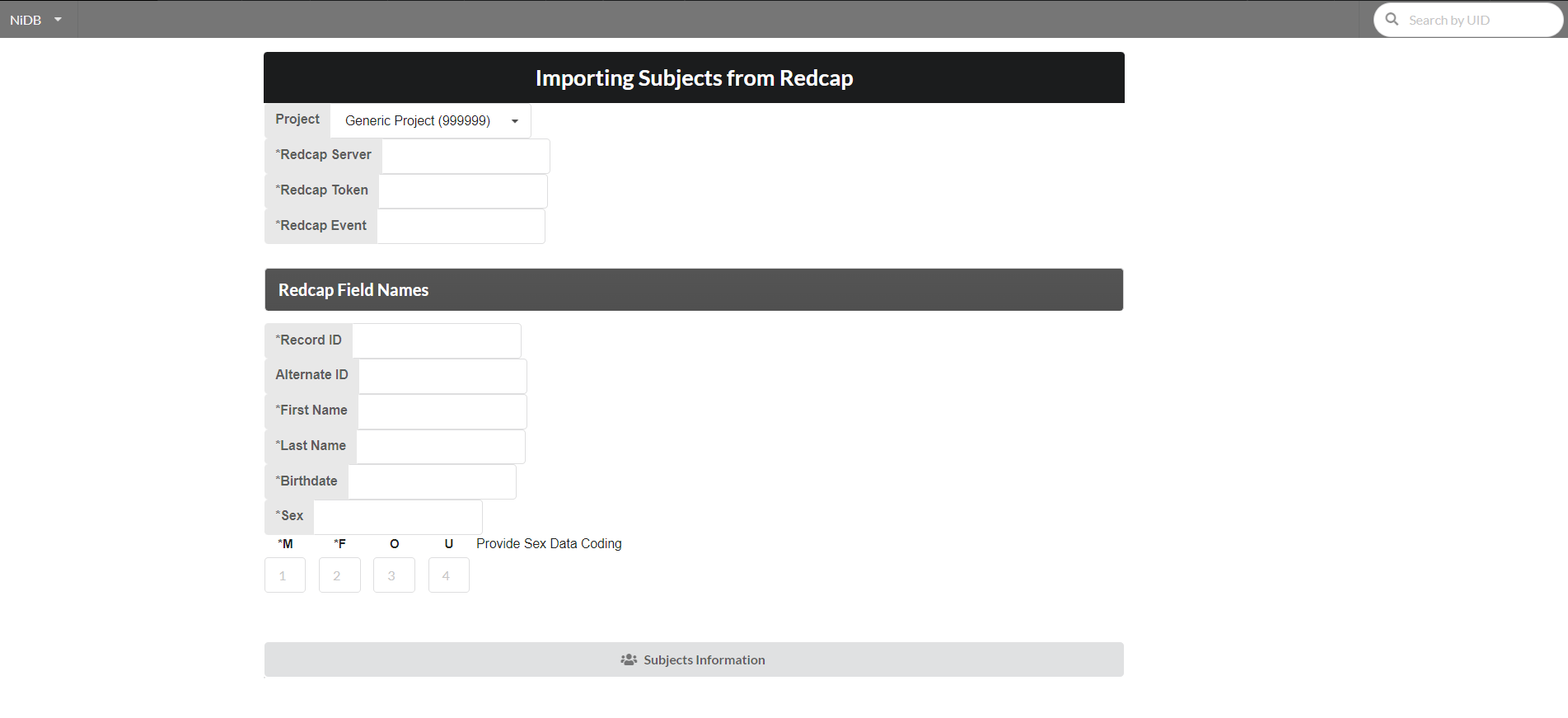
If all the above information is correct, then the list of the subjects from redcap will be shown as follows:
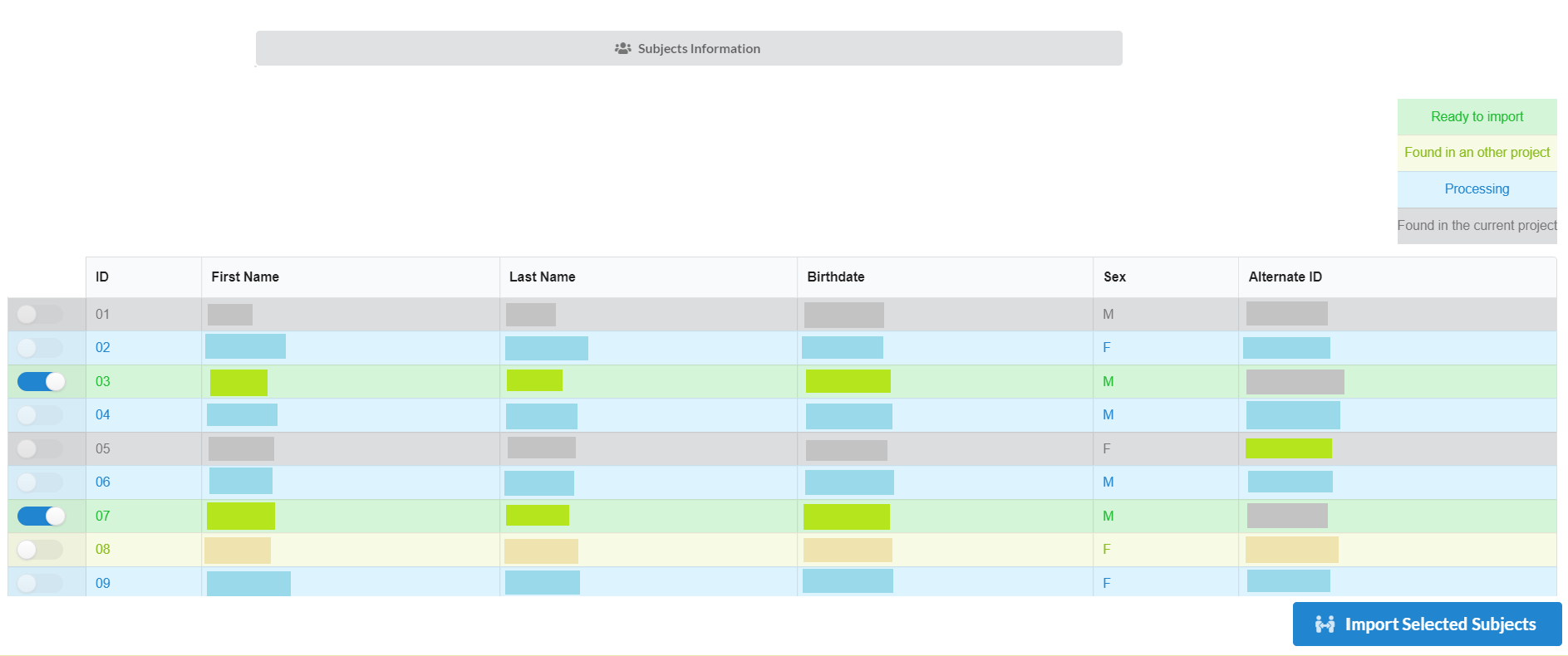
There can be four types of subjects in the list. Those are:
Ready to Import: are the one those are in redcap and can be imported.
Found in an other project: these are present in another project under inthe NiDB database. They can also be imported, but need to be selected to get import.
Processing: these are already in the process of being imported and cannot be selected to import.
Already exist in the project: these already exist in the current project and cannot be duplicated.
After selecting the required subjects click "Import Selected Subjects" to start the import process.
Enroll Subject in a Project
In the enrollments section, select the project you want to enroll in, and click Enroll. The subject will now be enrolled in the project. Permissions within NiDB are determined by the project, which is in theory associated with an IRB approved protocol. If a subject is not enrolled in a project, the default is to have no permissions to view or edit the subject. Now that the subject is part of a project, you will have permissions to edit the subject's details. Once enrolled, you can edit the enrollment details and create studies.
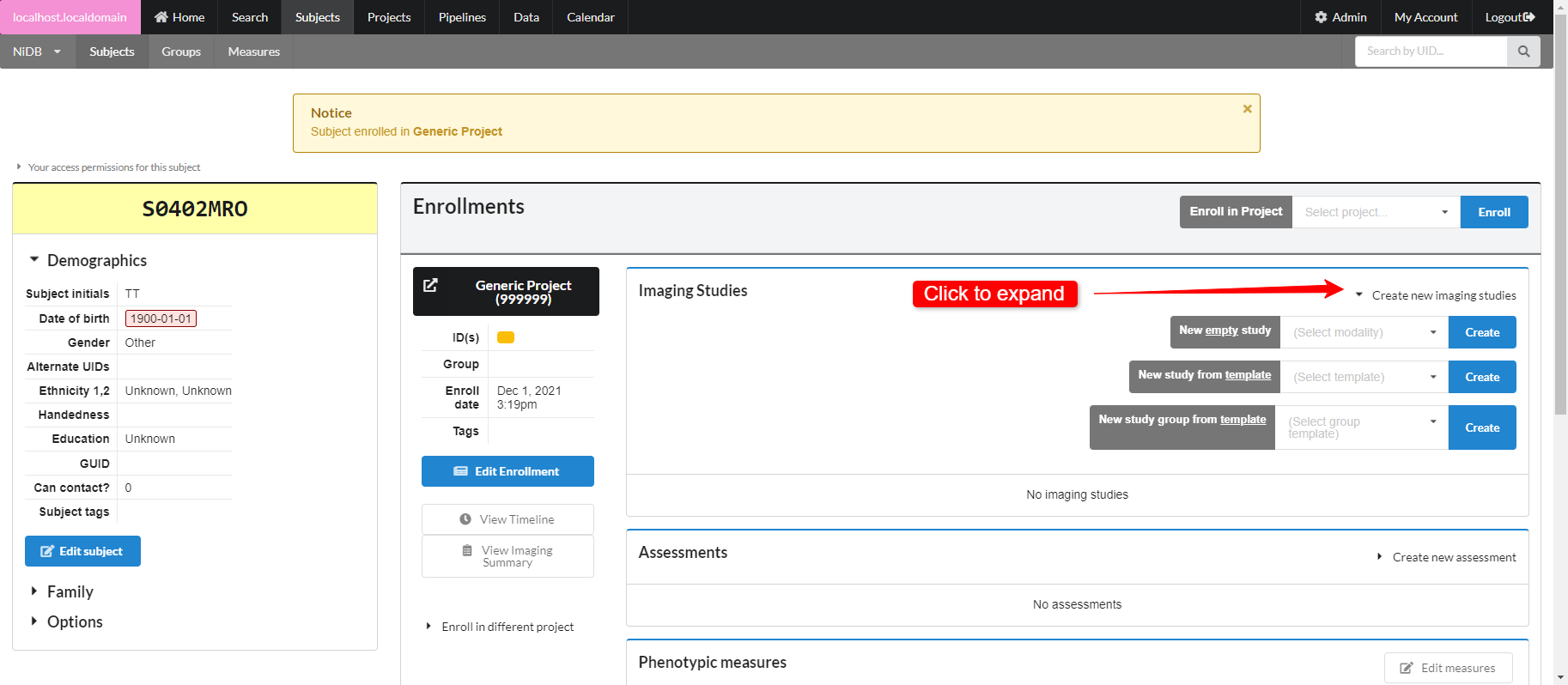
Create Imaging Study
There are three options for creating studies
Create a single empty study for a specific modality
Create a single study prefilled with empty series, from a template
Create a group of studies with empty series, from a template
Click Create new imaging studies to see these options. To create study templates or project templates, see Study Templates.
Once the study is created, it will appear in the list of imaging studies. Studies are given a unique number starting at 1 in order in which they are created. The studies are sorted by date in this list. While studies will often appear sequential by date and study number, this is because study numbers are incremented by each new study date added, and each new study often occurs at a later date. However, studies may be numbered in any order, regardless of date. If you create several studies for previous dates, if importing older data, if deleting or merging studies, this will cause study numbers to appear random. This is the normal behavior.
Create Single Series/Upload data
MRI and non-MRI data are handled differently, because of the substantial amount of information contained in MRI headers. MRI series are created automatically during import, while all other imaging data can be imported automatically or manually.
MRI
MRI series cannot be created manually, they must be imported as part of a dataset. See Bulk Import of Large Datasets or Automatic Import via DICOM receiver. MRI series can be managed individually after automatic importing has occured.
Non-MRI
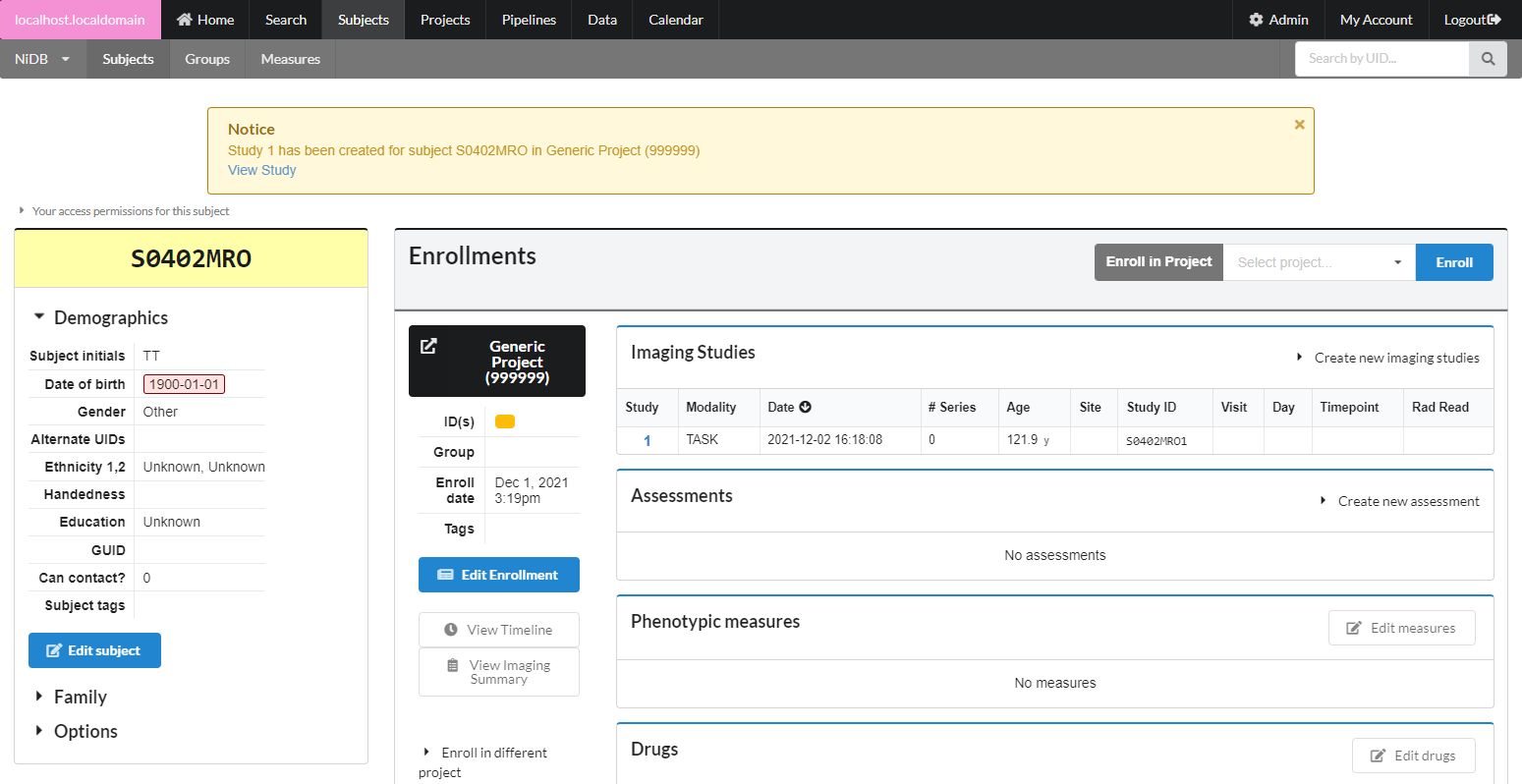
Non-MRI data be imported automatically or manually. To manually import non-MRI data, first go into the imaging study. Then fill out the series number, protocol, date, notes. Series number and date are automatically filled, so change these if you need to. When done filling out the fields, click Create Series.
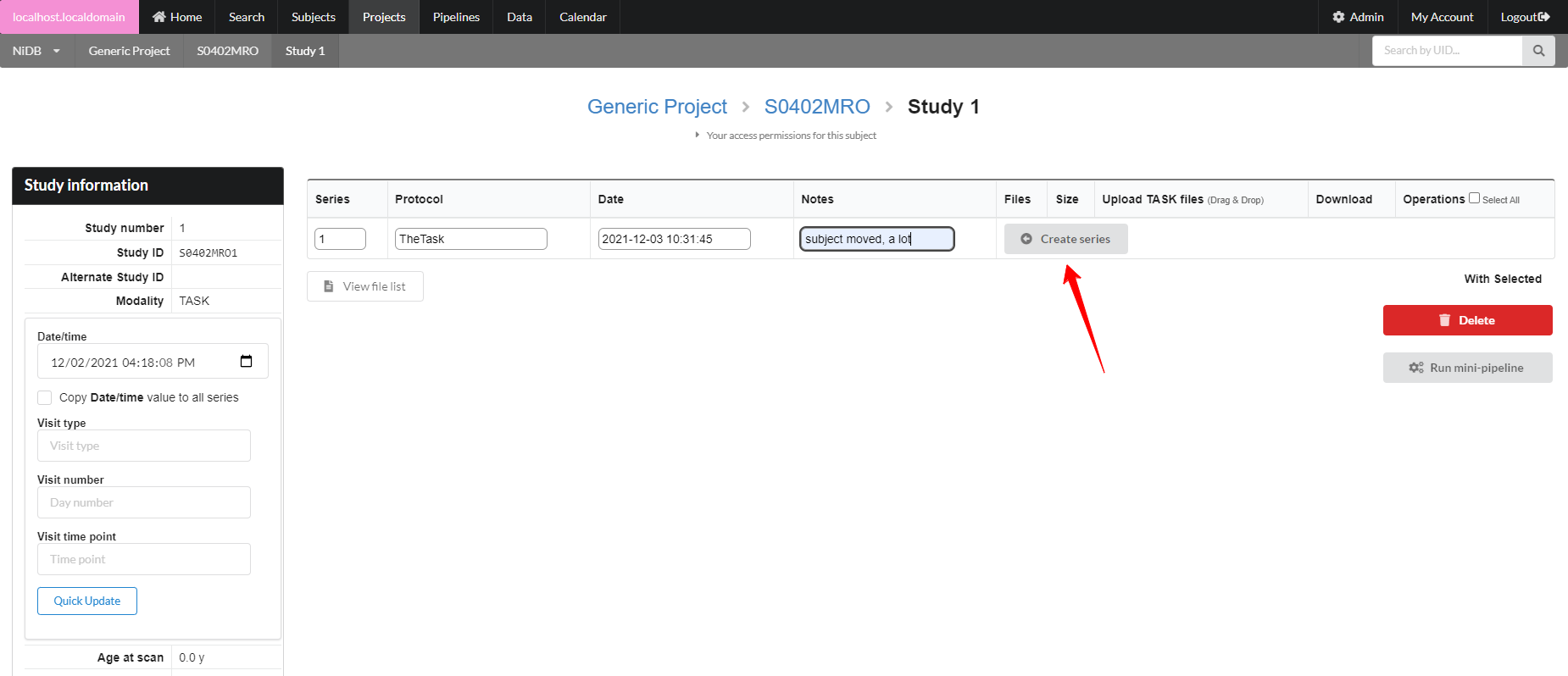
The series will be created, with an option to create another series below it. Upload files by clicking the Upload button, or by dragging and dropping onto the Upload button. If you need to delete or rename files, click the Manage files button. This will display a list of files in that series, and you can rename the file by typing in the filename box.
Bulk Import of Large Datasets
The imaging import page can be accessed by the Data → Import Imaging menu. Because datasets can be large and take hours to days to completely import and archive, they are queued in import jobs. To import a dataset, click the New Import button.
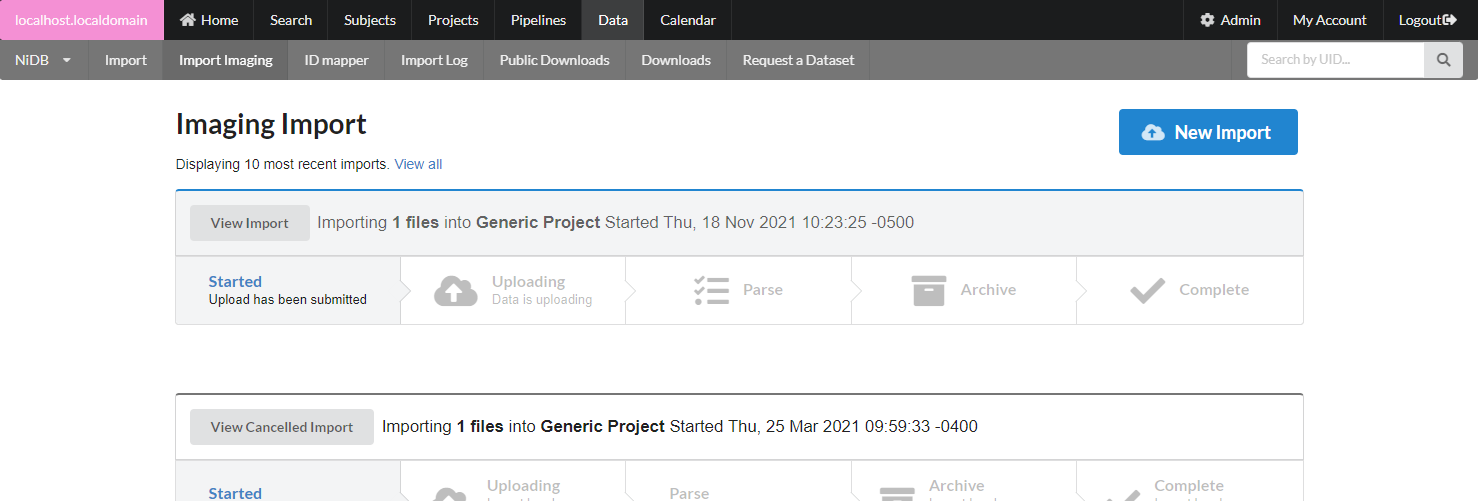
This will bring up the new import page.
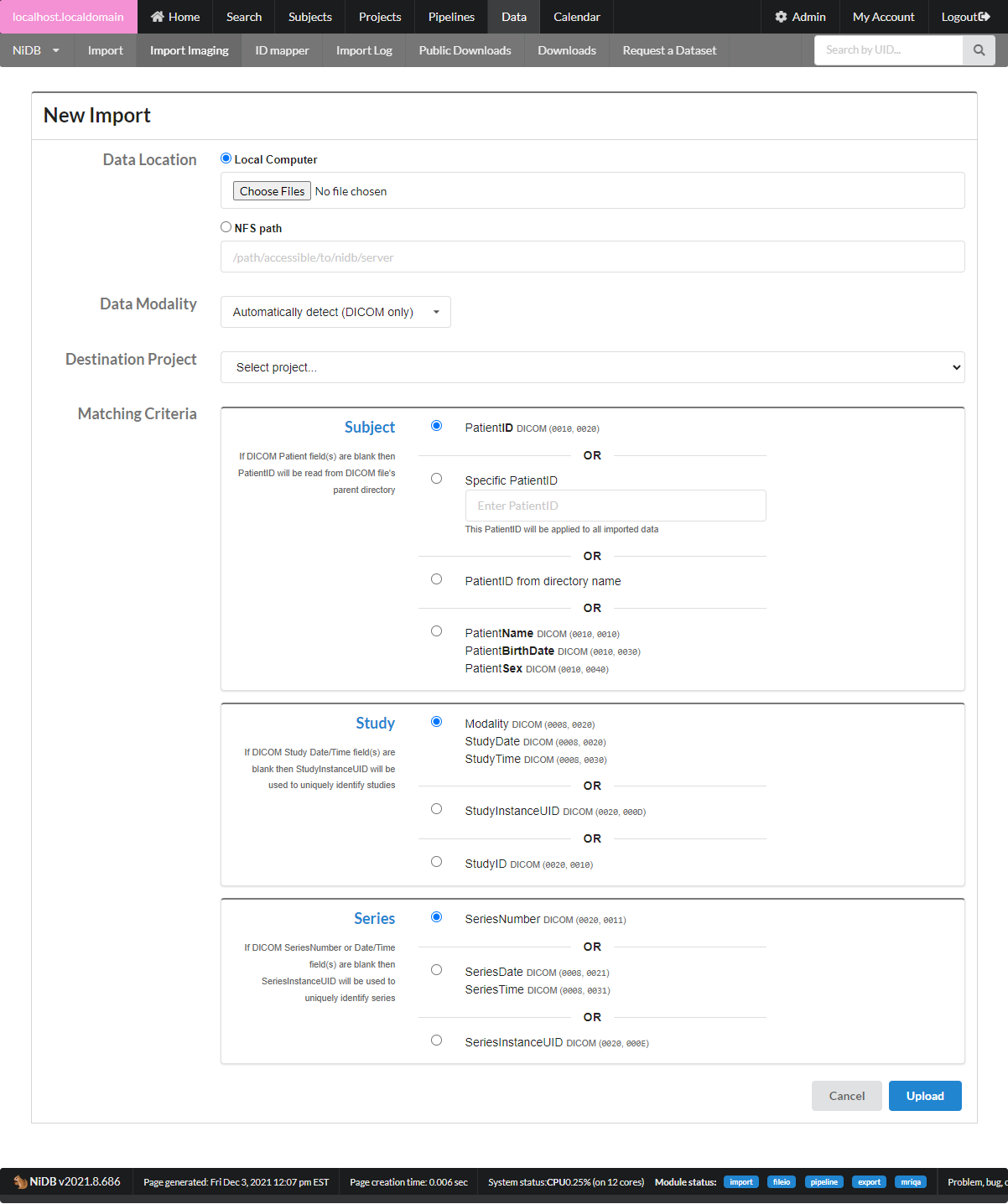
Data Location
Local computer
Upload files via the web browser. 'Local computer' is basically the computer from which the browser is being run, so this may be a Windows PC, Mac, or other browser based computer
NFS path
This is a path accessible from NiDB. The NiDB admin will need to configure access to NFS shares
Data Modality
Automatically detect
This option will detect data modality based on the DICOM header. If you are importing DICOM data, use this option
Specific modality
If you definitely know the data being imported is all of one modality, chose this. Non-DICOM files are not guaranteed to have any identifying information, so the imported files must be named to encode the information in the name.
Unknown
This is a last ditch option to attempt to figure out the modality of the data by filename extension. It probably won't work
Destination Project - Data must be imported into an existing project.
Matching Criteria - DICOM data only
Subject
PatientID - match the DICOM PatientID field to an existing UID or alternate UID
Specific PatientID - this ID will be applied to all imported data, ex S0001 will be the ID used for all data in the entire import
PatientID from directory name - get the subject ID from the parent directory of the DICOM file. This will be the highest level directory name, ex for 12345/1/data/MRI the subject ID will be 12345
Study
Default is to match studies by the DICOM fields Modality/StudyDate/StudyTime. Sometimes anonymized DICOM files have these fields blank, so StudyInstanceUID or StudyID must be used instead. If data is not importing as expected, check your DICOM tags and see if these study tags are valid
Series
The default is to match series by the DICOM field SeriesNumber. But sometimes this field is blank, and SeriesDate/SeriesTime or SeriesUID must be used instead. If data is not importing as expected, check your DICOM tags to see if these series tags are valid
After all of the import information is filled out, click Upload. You can view the import by clicking on it. The import has 5 stages, described below.
Started
The upload has been submitted. You will likely see this status if you are importing data via NFS, rather than through local web upload
Upload
Uploading - The data is being uploaded Uploaded - Data has finished uploading
Parsing
Parsing - The data is being parsed. Depending on the size of the dataset, this could be minutes, hours, or days Parsed - The data has been parsed, meaning the IDs, series, and other information have been read and the data organized into a Subject→Study→Series heirarchy. Once parsing is complete, you must select the data to be archived
Archive
Archiving - The data is being archived. Depending on the size of the dataset, this could be minutes, hours, or days Archived - The data is finished archiving
Complete
The entire import process has finished
Once the parsing stage is complete, you will need to select which series you want to import. This step gives you the opportunity to see exactly what datasets were identified in the import. If you were expecting a dataset to be in the import, but it wasn't found, this is a chance to find out why. Parsing issues such as missing data or duplicate datasets are often related to the matching criteria options. Sometimes the uniquely identifying information is not contained in the DICOM field it is supposed to be. That can lead to all series being put into one subject, or a new subject/study created for each series. There are so many ways in which data is organized and uniquely identified, so careful inspection of your data headers is important to select the right options.
If you find that none of the available matching options work for your data, contact the NiDB development team because we want to cover all import formats!
After you've selected the series you want to archive, click the Archive button. This will move the import to the next stage and queue the data to be archived.
At the end of archiving, the import should have a complete status. If there are any errors, the import will be marked error and you can view the error messages.
Automatic Import via DICOM receiver
NiDB was originally designed to automatically import MRI data as it is collected on the scanner, so this method of import is the most robust. After each series is reconstructed on the MRI, it is automatically sent to a DICOM node (DICOM receiver running on NiDB). From there, NiDB parses incoming data and will automatically create the subject/enrollment/study/series for each DICOM file it receives.
How to make DICOM imports more efficient
Write mosaic images - Depending on the MRI scanner, the option to write one DICOM file per slice or per volume may be available. On Siemens MRIs, there should be an option for EPI data to write mosaic images. For example, if your EPI volume has 36 slices, the scanner would normally write out 36 separate files, each with an entire DICOM header. If you select write mosaic images, it will write one DICOM file with one header for all 36 slices. If you have 1000 BOLD reps in a timeseries, this time savings can be significant.
Ignore phase encoding direction - To read the phase encoding direction information from a Siemens DICOM file can require 3 passes to read the file, using 3 different parsers. Siemens contain a special section called the CSA header which contains information about phase encoding direction, and an ASCII text section which includes another phase encoding element, and the regular DICOM header information. Disabling the parsing of phase encoding direction can significantly speed up the archiving of DICOM files.
Bulk Upload of non-MRI data
For non-MRI data, you can upload data in bulk to existing series. For example, if you have a directory full of task files, but each file belongs to a different subject. Rather than go into each subject/study and upload the file individually, you can upload the files as a batch. This method is best when used in conjunction with study templates.
This upload method assumes that you have already created all of the subjects, studies, and series. The series can be empty, or not. To create empty studies by template, see the Create Imaging Study section on use of templates.
Start by searching on the Search page for the series you are interested in uploading data into. For example, search for all 'GoNoGo' TASKs in a particular project. This will show a list of just the series from that project, from the TASK modality, and for existing GoNoGo series. Select the series you want, and go toward the bottom of the page, in the Operations section, click the Batch Upload button.
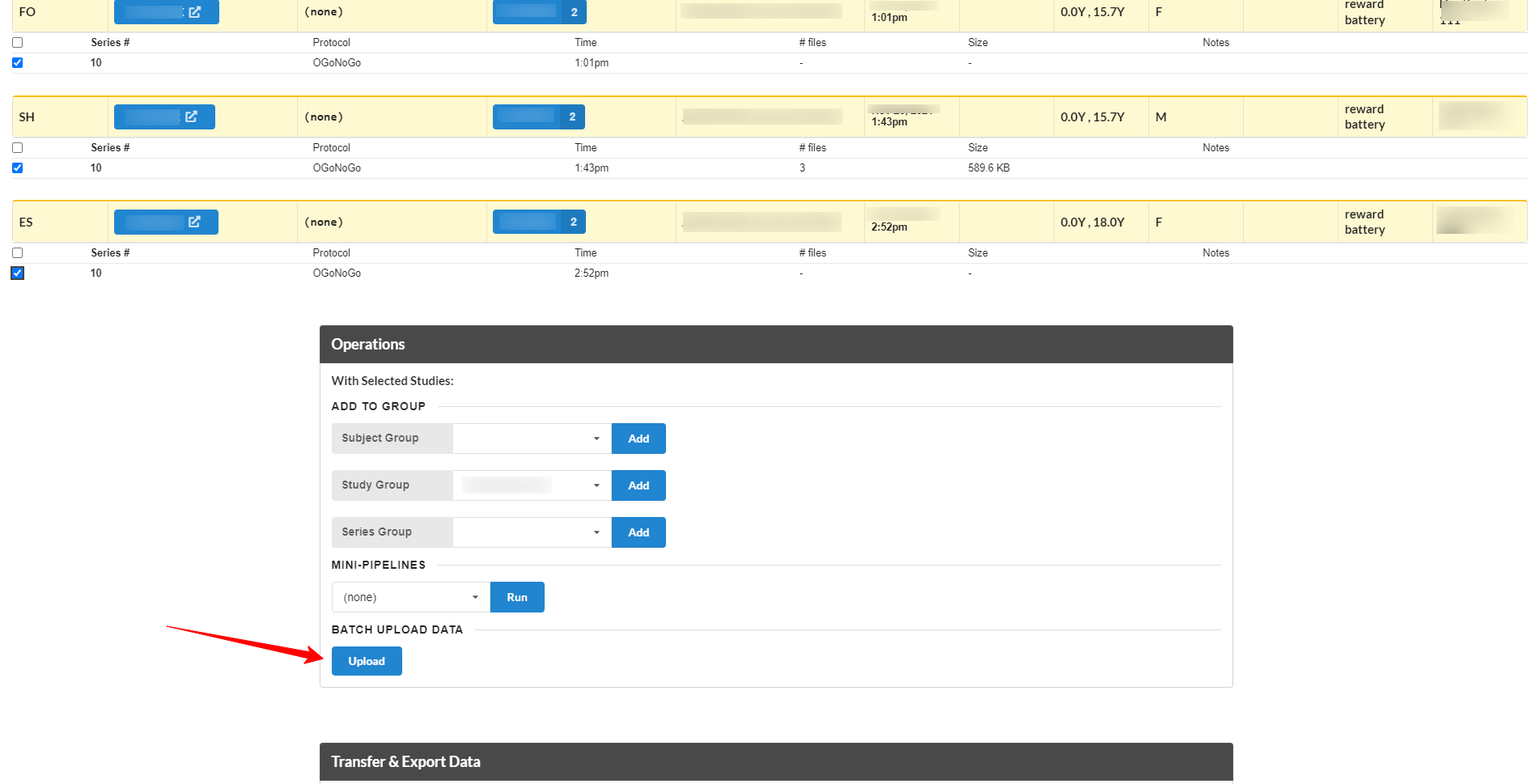
This will display a list of just those series, with an area to drag&drop files onto. Existing files for each series are displayed on the right side of the page.
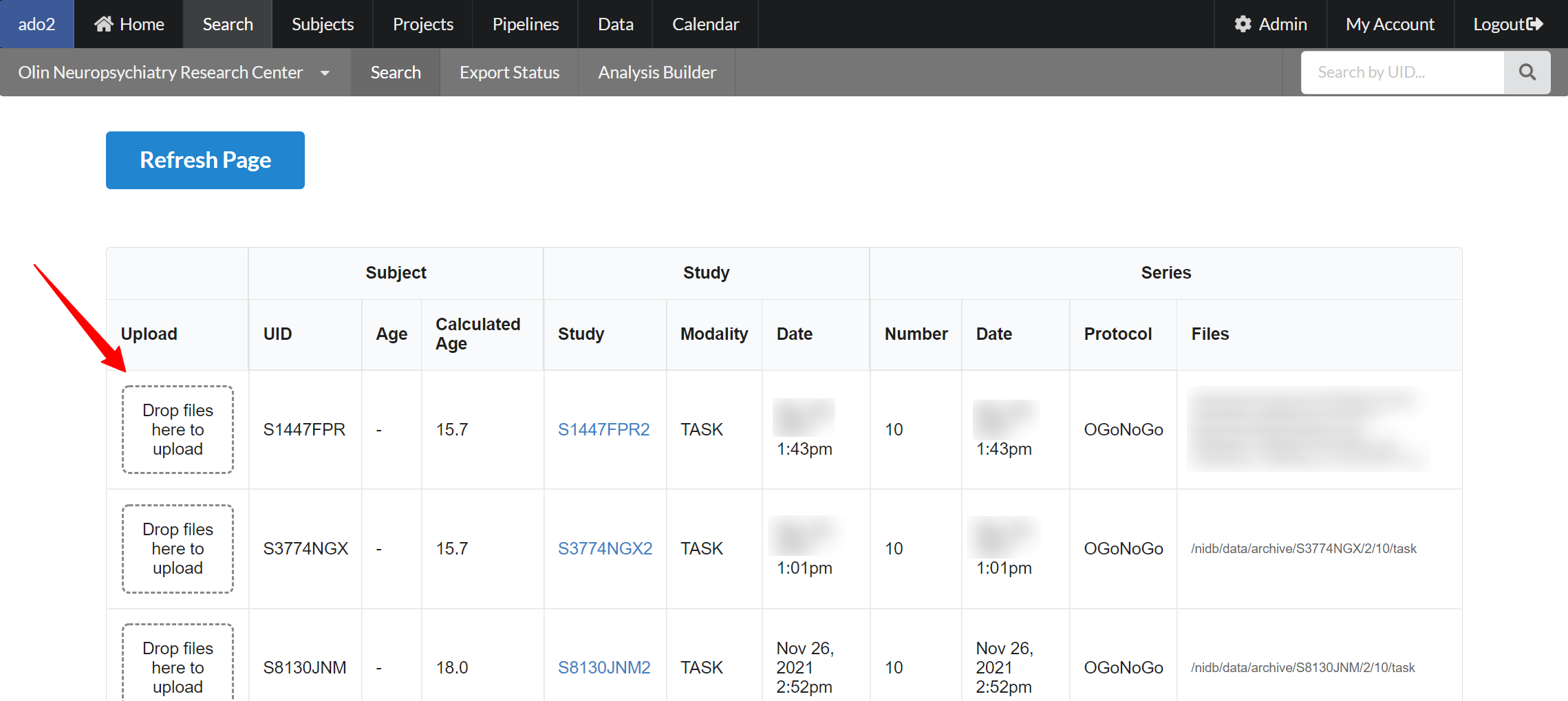
Drag and drop files onto those series, and click Refresh Page to view the newly uploaded files.
Last updated
Was this helpful?
